| Name | SHINTANI Takahiro |
|---|---|
| Position | Professor |
| Tel | 022-757-4445 |
| takahiro.shintani.d7*tohoku.ac.jp(Please replace * with @) | |
| Research Interest | Protein metabolism of eukaryotic microorganisms (yeast and koji mold), production of useful substances using eukaryotic microorganisms |
| Career | Education: B.S. Faculty of Agriculture, Tohoku Univ. (1991), PhD (Agri.)(1997) Research Experience: Research Fellowships for Young Scientists (DC2) (1996-1998 @ Tohoku Univ.), Research Fellowships for Young Scientists (PD) (1998-2001 @ National Institute for Basic Biology), Postdoctoral research fellow (2001-2004 @ Univ. of Michigan), Associate Prof., (2004-2020 @ Tohoku Univ), Prof., (2020- @ Tohoku Univ.) |
| Research map | https://researchmap.jp/read0116692?lang=ja |
| Research Projects | |
| |
| Research Seeds | |
The yeast Saccharomyces cerevisiae has been used for basic research as a model organism for eukaryotes and has long been used for alcoholic fermentation around the world. S. cerevisiae is also an unusual creature in that it ferments alcohol in the presence of glucose and other fermentable carbon sources (in nature, fruits, tree sap and flower nectar) even in the presence of oxygen. Even if other carbon sources are available, the fermentable carbon source is preferentially consumed. When the fermentable carbon source in the environment is exhausted, the energy production shifts from glycolytic-fermentative to respiration-based energy production, and the gene expression changes drastically. Jen1 is a lactate/pyruvate transporter localized in the plasma membrane, and its expression is significantly activated by the conversion from fermentation to respiration (i.e., glucose depletion). However, when a fermentable carbon source is replenished, Jen1 on the plasma membrane is rapidly degraded. In our laboratory, we have clarified the mechanism by which Jen1 is selectively degraded in response to glucose. When respiring yeast cells are exposed to glucose, the C-terminal region of Jen1 is recognized and ubiquitinated by the Rod1-Rsp5 ubiquitin ligase complex, leading to its degradation in the vacuole via endocytosis. When this C-terminal region (20 amino acid residues) was fused to another differentially regulated transporter (methionine transporter), it was degraded in a glucose- and Rod1-dependent manner. In other words, this region is a Rod1-dependent glucose-responsive degron (degradation control region). These results are expected to contribute not only to the elucidation of the regulation of nutrient uptake in response to changes in nutrient sources in S. cerevisiae, but also to the elucidation of the regulation of sugar uptake in mammals. 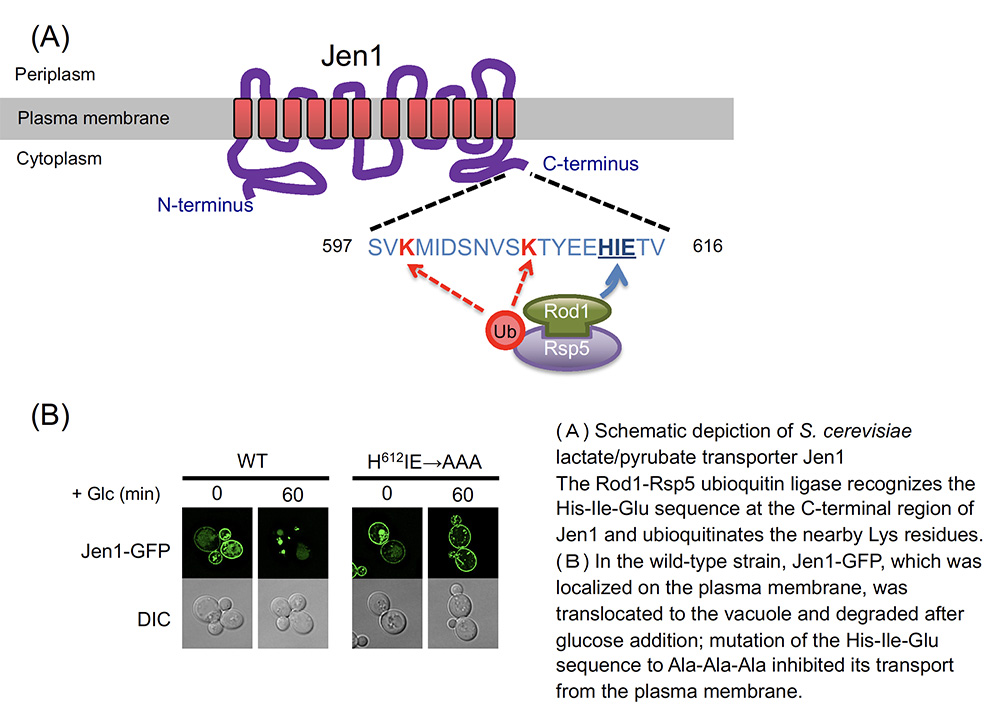 |
|



