
Development of breeding techniques using SNP analysis of genes
In breeding, gselectionh of individuals or lines having desirable genotypes is the most important step. However, it is difficult to select desirable genotypes of ingredient traits, disease/pest resistances, or some physiological traits by evaluation of phenotypes. Selection of desirable genotypes by DNA analysis is expected to realize efficient and reliable selection based on genome information. Since there can be differences of only one nucleotide between a desirable allele and an undesirable allele, DNA analysis used in breeding is required to enable detection of single nucleotide polymorphism (SNP) in any nucleotide sequences. Therefore, in our laboratory, we have developed simple and cost-effective SNP analysis techniques. Among various SNP analysis techniques including our techniques, the dot-blot-SNP technique developed in our laboratory is considered to be the most cost-effective and laborsaving technique applicable to analysis of a large number of individuals, and, therefore, is expected to be useful in practical breeding.
1. Improvement of techniques for detection of unidentified SNPs using electrophoresis
Nucleotide sequencing of genes, sequence differences of which are unknown, is generally performed for identification of SNPs, but it is costly. Electrophoresis of DNA enables detection of SNPs of many samples with low cost.
We first adopted the SSCP (single strand conformation polymorphism) method, which has been developed by Japanese scientists, for analysis of SNPs. This method is simple and cost-effective, but it was sometimes difficult to detect SNPs in fragments longer than 500 bp and those at the end of DNA fragments. A modified PCR-RF-SSCP method, in which PCR (polymerase chain reaction) products of ca. 2 kb cleaved by several four-base cutters of restriction endonuclease are subjected to SSCP analysis, was found to be useful for efficient SNP detection. Using this method, we identified many SNPs of mutations in rice wx gene (Sato & Nishio 2003 Theor Appl Genet 107, 560-567). This method was also applied to detection of SNPs in many genes of rice cultivars (Shirasawa et al. 2004 DNA Res 11, 275-283).
Mutant selection from chemically mutagenized populations using an SNP analysis method, in which mismatch sites of double strand DNA fragments are cleaved by heteroduplex cleavage nuclease extracted from celery, named TILLING, is widely used in the world. In this laboratory, we used petioles of Mizuna (Brassica rapa) instead of celery because Mizuna petioles are easily pulverized in liquid nitrogen, and enabled selection of rice mutants from mutagenized populations by gamma-rays (Sato et al. 2006 Breeding Sci 56, 179-183).
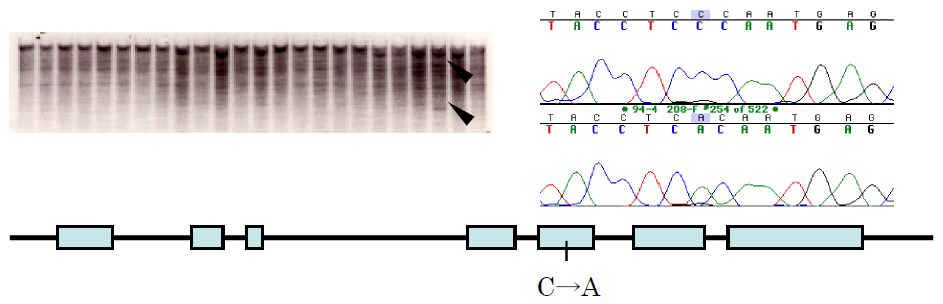
Figure. Detection of mutations induced by gamma-rays
2. Development of the dot-blot-SNP technique
Electrophoresis for SNP analysis is laborious, in which one person can analyze less than 300 samples per day, and is not suitable for analysis of a large number of samples or genes. Therefore, we developed the dot-blot-SNP technique, which enables SNP genotyping of a large number of individuals (Shirasawa et al. 2006 Theor Appl Genet 113, 147-155). In this method, DNA fragments amplified by PCR are dot-blotted on a nylon membrane, and hybridized with a labeled 17-mer oligonucleotide together with an unlabeled oligonucleotide having the sequence of the other allele. Similar method has already been reported, but it has not been used because of high background signals. Our method is an improved method, in which problem of high background signal was solved.

Figure. Using this machine, 864 DNA samples can be dot-blotted onto nylon membrane of 8 mm ~ 12 mm.
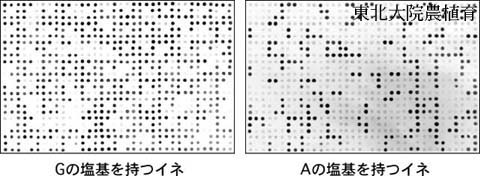
Figure. SNP genotyping of plants in rice breeding program by dot-blot-SNP.
SNP genotyping of 864 individuals can be performed using two sheets of nylon membrane in dot-blot-SNP, and 10 to 20 sheets can be handled at a time by one person, enabling SNP genotyping of ca. 4,000 to 8,000 individuals. A large number of plants handled in conventional plant breeding can be analyzed with low cost by this method. When a large number of plants are analyzed, DNA preparation from plants for PCR is laborious. Since short DNA fragments amplified by PCR can be used for hybridization in dot-blot-SNP, a small leaf piece cut by a mini cork borer was found to be usable as a PCR template (Shiokai et al. 2009 Mol Breed 23, 329-336).
Although dot-blot-SNP is suitable for analysis of a large number of individuals, analysis with many SNP sites is costly, because it requires synthesis of labeled oligonucleotide probes. To reduce the cost for oligonucleotide labeling, we also developed an improved method using indirect labeling of oligonucleotide probes (Shiokai et al. 2010 Mol Breed 25, 179-186). Hybridization conditions should be optimized for each SNP marker, and, therefore, SNP analyses using many SNP markers are laborious. We developed a method for estimation of optimum hybridization and washing conditions (Shiokai et al. 2010 Plant Cell Rep 29, 829-834).
Using the improved dot-blot-SNP technique, we constructed linkage maps of genes having SNPs in B. rapa, B. oleracea, and R. sativus, and analyzed QTLs for various traits in rice and Brassicaceae crops (Please see gStudies on genome and genetic resources in Brassicaceae cropsh, gMolecular genetic study on plant reproduction mechnismsh and gStudies on genetics and breeding of environmental stress tolerancesh). Cooperative research with the National Agricultural Research Center for Tohoku Region revealed 18 dot-blot-SNP markers are sufficient for identification of one rice cultivar among ca. 200 cultivars grown in > 10 ha fields in Japan (Sato et al. Breeding Sci 60, 447-451).
Since the dot-blot-SNP technique requires long time as Southern blot analysis (usually 24 hours), it is not suitable for analysis of a small number of individuals. If temperature for hybridization and washing is not correct, reliable signals cannot be obtained. Therefore, we developed a method much easier than dot-blot-SNP. The principle of this method is the same as that of dot-blot-SNP, but streptavidin-coated magnetic beads are used for fixation of DNA fragments instead of nylon membrane. Hybridization and signal detection can be carried out in one PCR machine, and, therefore, temperature setting is easy and precise. Results can be obtained within several hours.
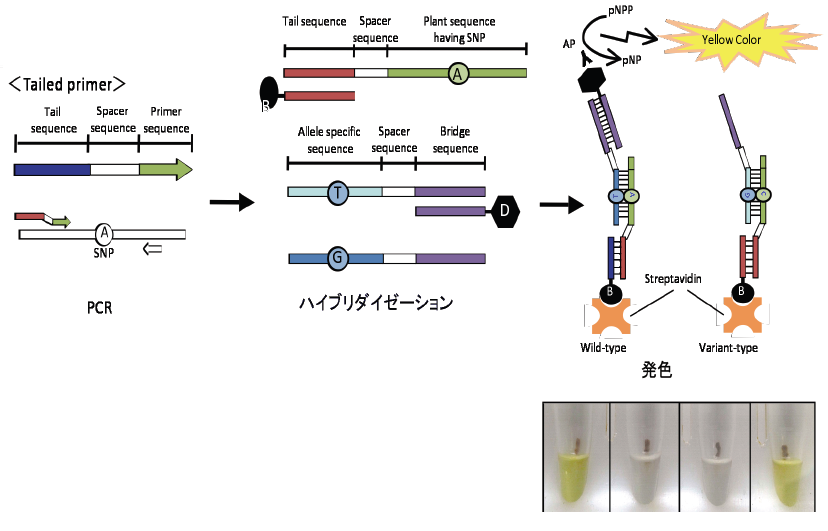
Figure. Scheme of magnetic bead hybridization for SNP analysis
A problem of SNP analysis using streptavidin-coated magnetic beads is high cost of the beads. Although this method is inferior to dot-blot-SNP in its cost, it is much easier and convenient. This method is useful not only in SNP analysis but also in species identification of plants and identification of S haplotypes in Brassicaceae and Rosaceae (Tonosaki et al. 2013 Mol Breed 31, 419-428; Wang et al. 2013 Plant Cell Rep 32, 567-576).